
Using Trials
trials.RmdIn many situations, behaviour is collected in trials. For this
reason, we have provided trials in wombats and
the align_trials parameter in
geom_ethogram().
Understanding Trials
Let’s explore the nature of the dataset. This is how it looks like when not visualizing trials.
ggplot(wombats,
aes(x=seconds,
y = wombat,
behaviour = behaviour,
color=behaviour)) +
geom_ethogram()
#> No observation interval provided, using guessed interval 5
Here’s when we facet by trial:
ggplot(wombats,
aes(x=seconds,
y = wombat,
behaviour = behaviour,
color = behaviour)) +
geom_ethogram() +
facet_wrap(~trial)
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5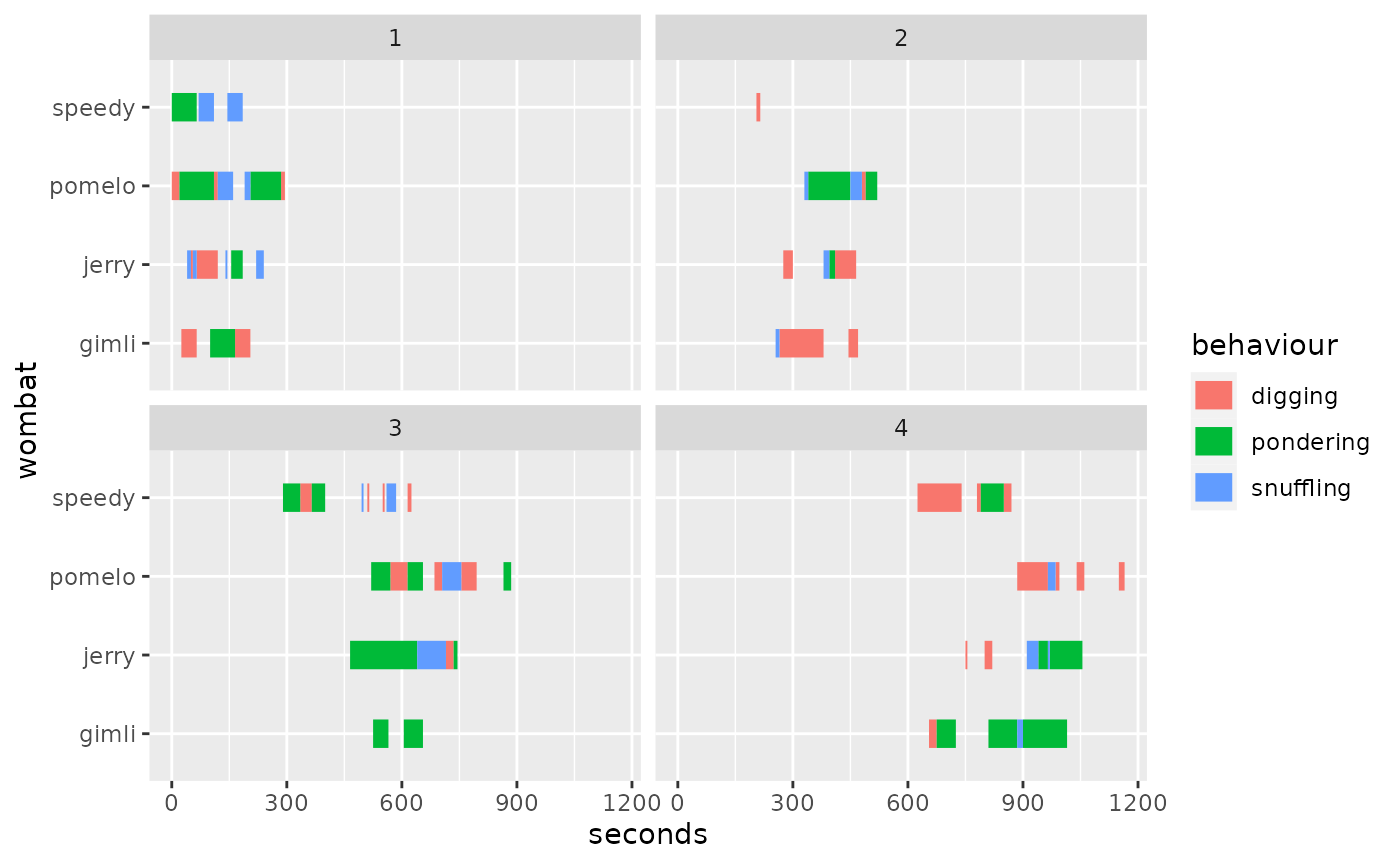
We can now see the issue: the trials were collected with a continuous
clock and we would like to align them to common time (i.e., have all
trials start at zero). This is when align_trials comes
handy:
ggplot(wombats,
aes(x=seconds,
y = wombat,
behaviour = behaviour,
color = behaviour)) +
geom_ethogram(align_trials=TRUE) +
facet_wrap(~trial)
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5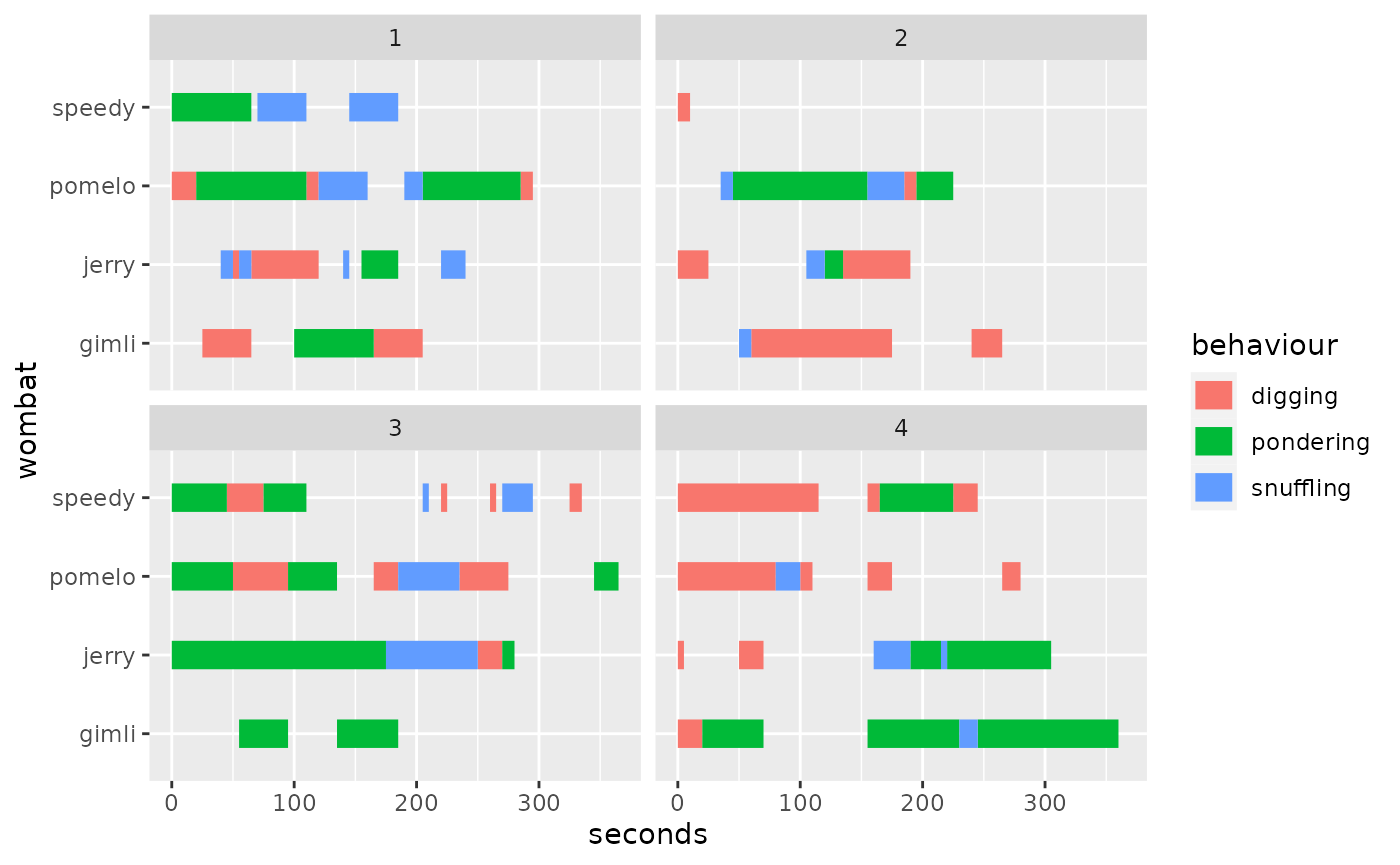
Here’s an another example. Let’s say we wanted to facet by
wombat and have trial be on the y
axis instead.
ggplot(wombats,
aes(x=seconds,
y = trial,
behaviour = behaviour,
color = behaviour)) +
geom_ethogram() +
facet_wrap(~wombat)
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> Warning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
#> dplyr 1.1.0.
#> ℹ Please use `reframe()` instead.
#> ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
#> always returns an ungrouped data frame and adjust accordingly.
#> ℹ The deprecated feature was likely used in the ggethos package.
#> Please report the issue to the authors.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> No observation interval provided, using guessed interval 5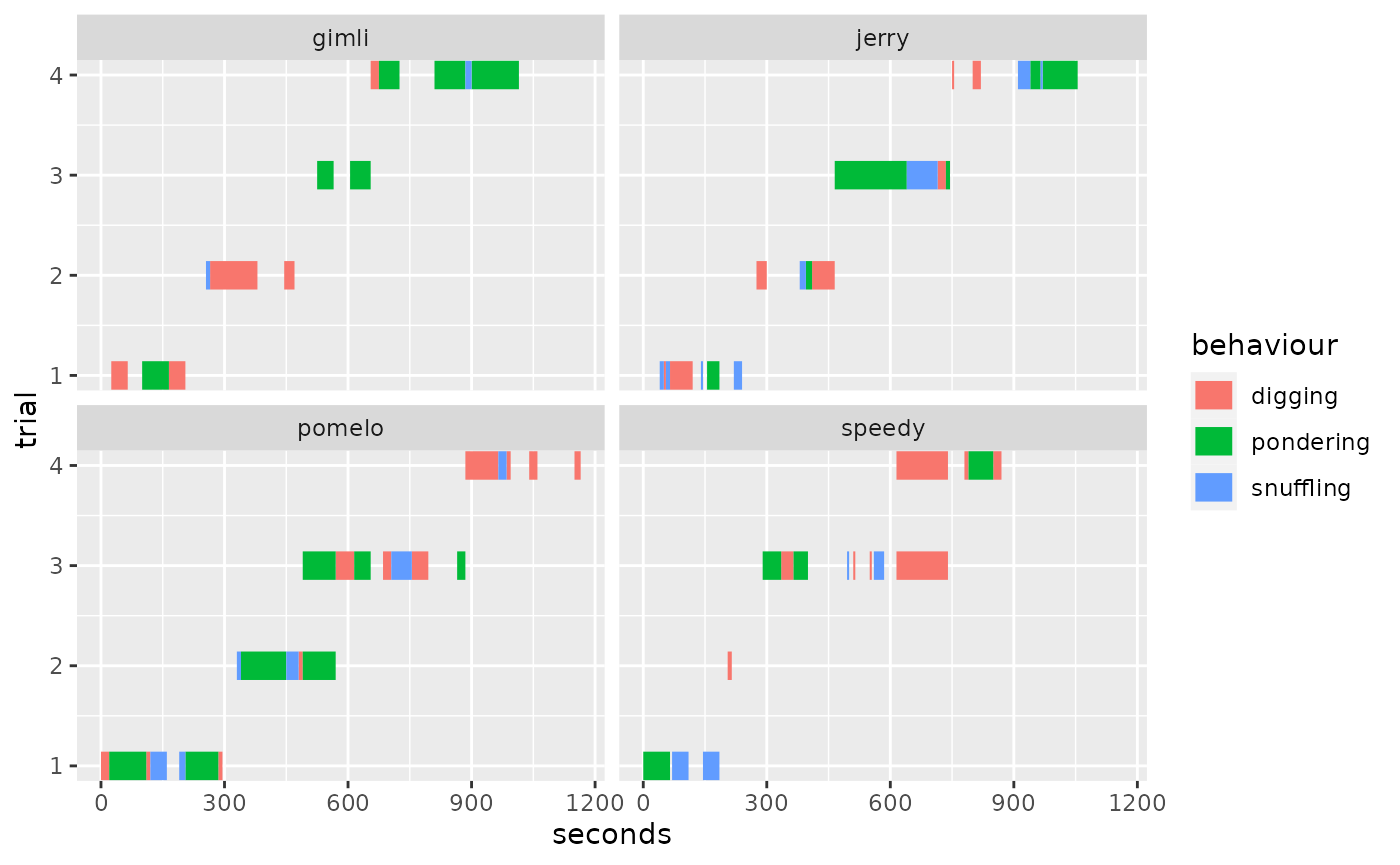 We can easily align trials again using
We can easily align trials again using align_trials=T:
ggplot(wombats,
aes(x=seconds,
y = trial,
behaviour = behaviour,
color = behaviour)) +
geom_ethogram(align_trials = T) +
facet_wrap(~wombat)
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5
#> No observation interval provided, using guessed interval 5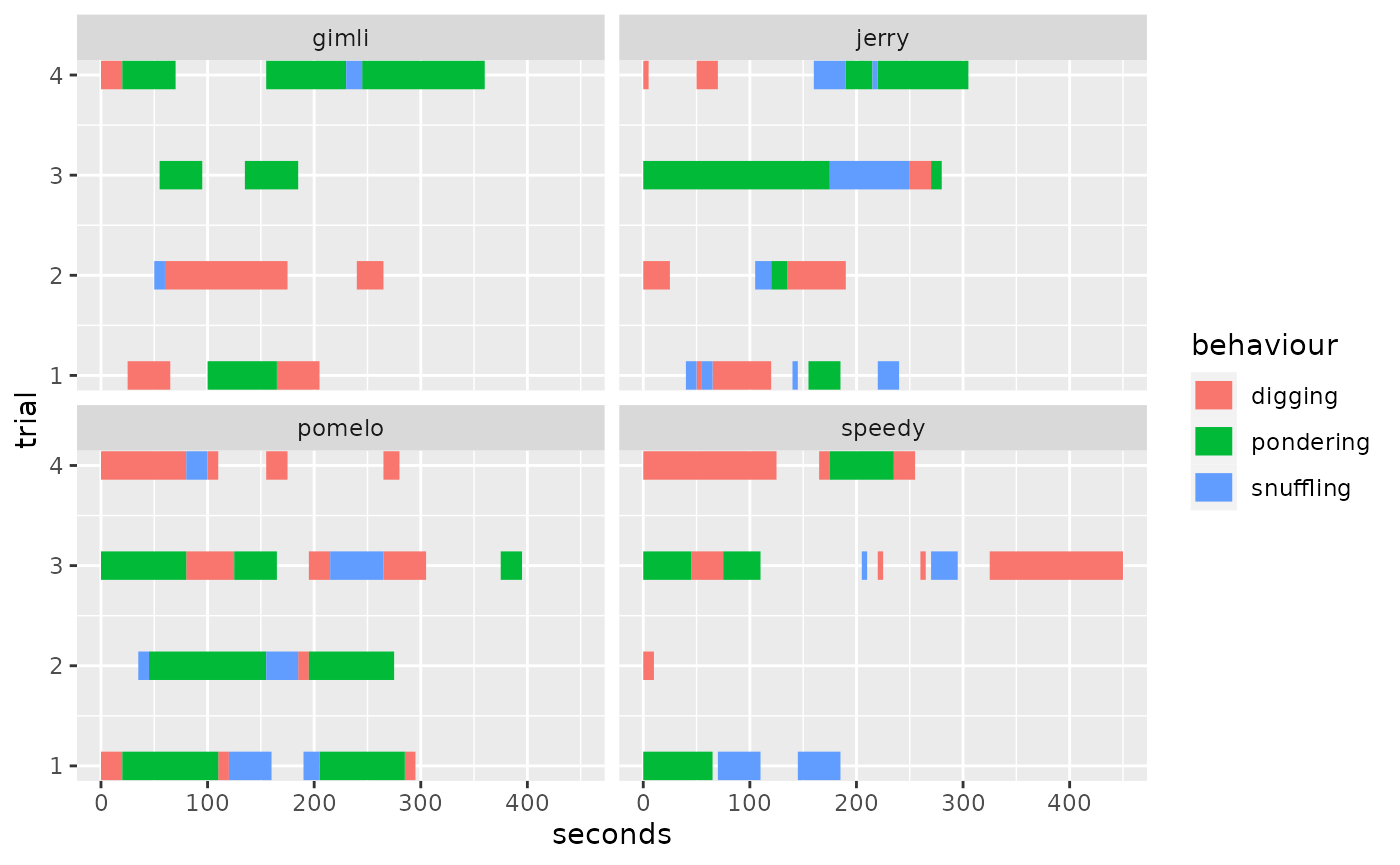
Summary
In this article, we have covered basic functionality of aligning
trials by using the align_trials argument.
Session Info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.2.3 (2023-03-15)
#> os Ubuntu 22.04.2 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate C.UTF-8
#> ctype C.UTF-8
#> tz UTC
#> date 2023-03-28
#> pandoc 2.19.2 @ /usr/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> bslib 0.4.2 2022-12-16 [1] RSPM
#> cachem 1.0.7 2023-02-24 [1] RSPM
#> cli 3.6.1 2023-03-23 [1] RSPM
#> colorspace 2.1-0 2023-01-23 [1] RSPM
#> crayon 1.5.2 2022-09-29 [1] RSPM
#> desc 1.4.2 2022-09-08 [1] RSPM
#> digest 0.6.31 2022-12-11 [1] RSPM
#> dplyr 1.1.1 2023-03-22 [1] RSPM
#> evaluate 0.20 2023-01-17 [1] RSPM
#> fansi 1.0.4 2023-01-22 [1] RSPM
#> farver 2.1.1 2022-07-06 [1] RSPM
#> fastmap 1.1.1 2023-02-24 [1] RSPM
#> fs 1.6.1 2023-02-06 [1] RSPM
#> generics 0.1.3 2022-07-05 [1] RSPM
#> ggethos * 0.0.0.9000 2023-03-28 [1] local
#> ggplot2 * 3.4.1 2023-02-10 [1] RSPM
#> glue 1.6.2 2022-02-24 [1] RSPM
#> gtable 0.3.3 2023-03-21 [1] RSPM
#> highr 0.10 2022-12-22 [1] RSPM
#> htmltools 0.5.5 2023-03-23 [1] RSPM
#> jquerylib 0.1.4 2021-04-26 [1] RSPM
#> jsonlite 1.8.4 2022-12-06 [1] RSPM
#> knitr 1.42 2023-01-25 [1] RSPM
#> labeling 0.4.2 2020-10-20 [1] RSPM
#> lifecycle 1.0.3 2022-10-07 [1] RSPM
#> magrittr 2.0.3 2022-03-30 [1] RSPM
#> memoise 2.0.1 2021-11-26 [1] RSPM
#> munsell 0.5.0 2018-06-12 [1] RSPM
#> pillar 1.9.0 2023-03-22 [1] RSPM
#> pkgconfig 2.0.3 2019-09-22 [1] RSPM
#> pkgdown 2.0.7 2022-12-14 [1] any (@2.0.7)
#> purrr 1.0.1 2023-01-10 [1] RSPM
#> R6 2.5.1 2021-08-19 [1] RSPM
#> ragg 1.2.5 2023-01-12 [1] RSPM
#> rlang 1.1.0 2023-03-14 [1] RSPM
#> rmarkdown 2.21 2023-03-26 [1] RSPM
#> rprojroot 2.0.3 2022-04-02 [1] RSPM
#> sass 0.4.5 2023-01-24 [1] RSPM
#> scales 1.2.1 2022-08-20 [1] RSPM
#> sessioninfo 1.2.2 2021-12-06 [1] any (@1.2.2)
#> stringi 1.7.12 2023-01-11 [1] RSPM
#> stringr 1.5.0 2022-12-02 [1] RSPM
#> systemfonts 1.0.4 2022-02-11 [1] RSPM
#> textshaping 0.3.6 2021-10-13 [1] RSPM
#> tibble 3.2.1 2023-03-20 [1] RSPM
#> tidyselect 1.2.0 2022-10-10 [1] RSPM
#> utf8 1.2.3 2023-01-31 [1] RSPM
#> vctrs 0.6.1 2023-03-22 [1] RSPM
#> withr 2.5.0 2022-03-03 [1] RSPM
#> xfun 0.38 2023-03-24 [1] RSPM
#> yaml 2.3.7 2023-01-23 [1] RSPM
#>
#> [1] /home/runner/work/_temp/Library
#> [2] /opt/R/4.2.3/lib/R/site-library
#> [3] /opt/R/4.2.3/lib/R/library
#>
#> ──────────────────────────────────────────────────────────────────────────────